Academic Genealogy
I compiled and designed an academic genealogy graphic and had it printed and framed as a gift. Here’s how I did it. Materials for you to do your own, including scripts and an example design, are available in this repository on GitHub. You’ll likely need some familiarity with Python syntax.
Supervisor Family Trees
Supervisor family trees are a fun bit of academic self-indulgence. These are like real family trees, but instead of depicting parent-child associations between individuals, a supervisor family tree depicts supervisor-student associations. Exactly what constitutes supervision is an open question – the most obvious definition is the supervision of a student’s doctoral thesis, but this is a bit too narrow. The current conception of a supervised, research-based PhD degree only originated in the 19th century, but the tradition of academic mentorship is far older (e.g., we could go back at least as far as Socrates and Plato). With a wider definition we can build an academic genealogy that goes back many centuries.
Having recently completed my own PhD, I took the opportunity to explore my own academic ancestry and put together a genealogy that I could give to my supervisors as a gift. (Also, by researching my supervisors' ancestry I’d also be researching my own – the perfect combination of self-indulgence and altruism!)
These academic family trees are nice because nearly all academics will be able to trace their ancestry back to at least a few notable scientists or mathematicians, in the same way that most western Europeans can trace their familial ancestry back to Charlemagne. Marin Mersenne, Isaac Newton, and Galileo Galilei are all ancestors of mine. In addition to direct ancestors, we can also look at individuals with whom one shares a common ancestor. For example, Alan Turing and Peter Hilton, both code-breakers at Bletchley Park during the Second World War, can be regarded as academic cousins as they both share Oswald Veblen as the supervisor of their respective doctoral supervisors.
The Data
The big challenge of compiling a genealogy is of course gathering the history of mentor-student relationships for those involved. Fortunately, the Mathematics Genealogy Project (MGP) has done a lot of the work for us. The MGP has mapped over 175,000 academics and their students. Although it is predominantly focused on mathematicians, it also includes academics who have made contributions in other fields, including physics, computer science, chemistry, and biology. A few people have written scripts and libraries that access this database to build a visualisation of an individual’s academic genealogy. The best I’ve found is David Alber’s Geneagrapher, which is written in Python. These scripts, however, only attempt to show an individual’s direct ancestors and descendants, not any interesting academics that they may share a common ancestor with.
Including common ancestors in the genealogy is a lot more challenging. The number of individuals that have a shared common ancestor with a typical living academic is going to be huge, resulting in a lot of queries to the MGP and producing an unwieldy visualisation. Instead, we want some way of selecting a few interesting individuals to see if their ancestry can be connected to the person we’re building a genealogy for (I’ll call this person the focal academic for short) and then building the visualisation around that, possibly culling a few unwanted branches of the tree in the process.
Some Scripts
The Script-GenealogyMiner directory contains a Python script, genealogy_miner.py, that crawls the MGP for a given focal academic, attempting to connect him/her to other academics. It’s configured through another Python file (specified as a command line argument) that contains configuration options. I’ve provided an example, config_turing.py, with Alan Turing as the focal academic. To run this example, download the Script-GenealogyMiner directory and execute:
python genealogy_miner.py config_turing.py turing.dot
For demonstration purposes, the configuration only has a few seed academics (see SEED_ID_LIST). Seeds are academics the script will attempt to find shared ancestry with. Crawling can take a while, depending on the number of individuals to be crawled, the number of ancestors they have, and the response time of the MGP servers. The crawl with the 14 example seed academics should take less than four minutes.
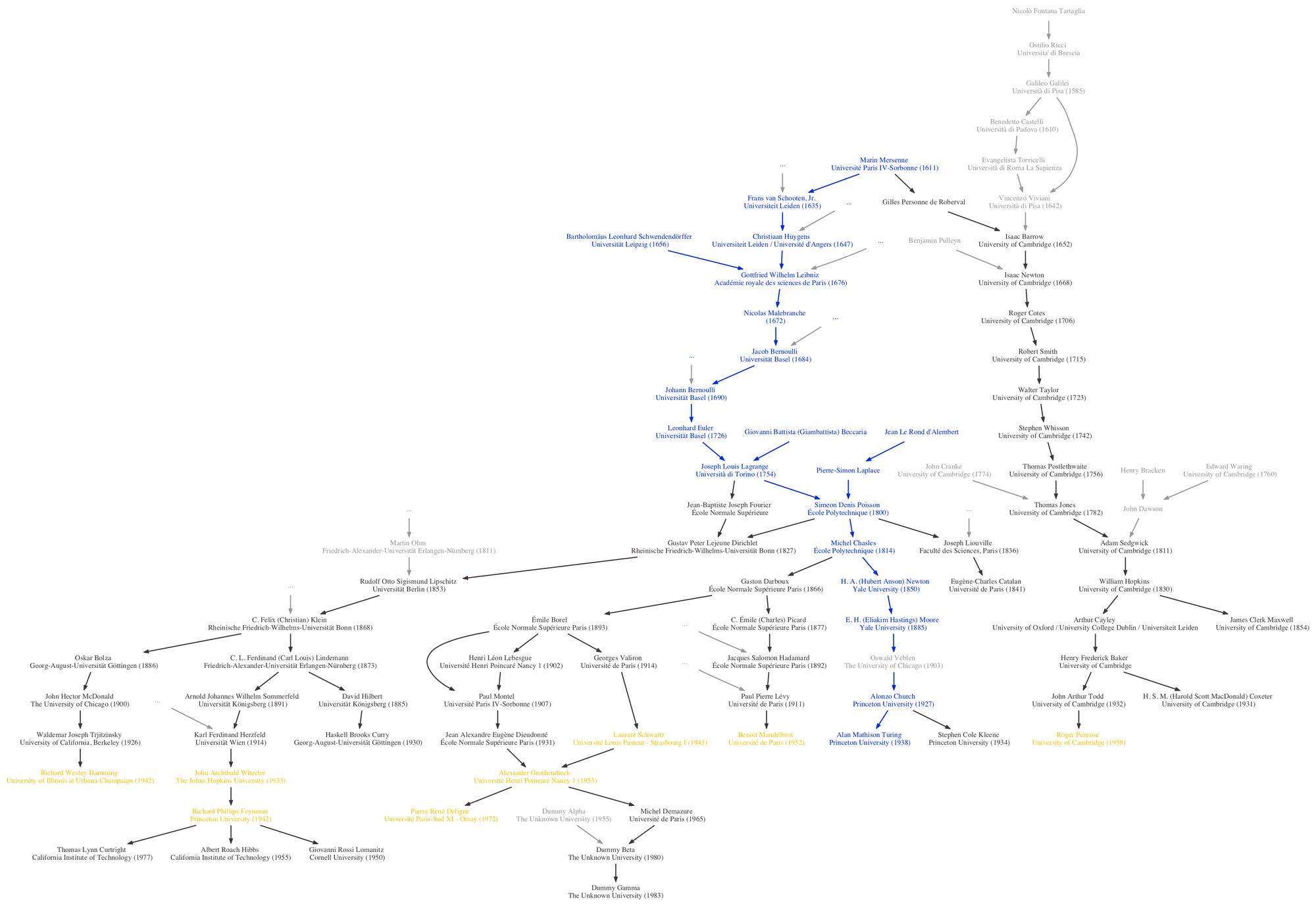
The output is a plain-text dot file (turing.dot) describing the genealogy (as a list of nodes and edges, including some formatting instructions such as text and arrow colours) that can be imported into other applications (I used OmniGraffle) so you can do further design work. dot is a popular graph description format and is fairly well supported. If you have
GraphViz installed on your system, you can have it generate a rendering via:
dot -T png turing.dot > turing.png
The GraphViz rendering isn’t production-quality – for the final graphic I imported the dot file into OmniGraffle – but it’s useful when you’re tweaking the crawl configuration. It takes a bit of guesswork to determine which academics might be reachable from the focal node. The configuration file allows you to specify a few different features which you’ll need to play around with, so having GraphViz on hand to do quick renderings of the resulting genealogy is useful.
I should note that the script builds on
Geneagrapher (already included in the Script-GenealogyMiner directory), which it uses to query the online MGP database.
Taking a look in config_turing.py shows how we can configure the crawler:
- The focal academic (
ID_FOCAL_NODE): The MGP identifier of the academic for whom we are generating the genealogy. This is given in the Mathematics Genealogy Project URL for a particular academic; e.g., Turing’s page ends in...?id=8014. - Prospective connections (
SEED_ID_LIST): A list of individuals in the MGP, again identified by their MGP identifier. The script will try to find common ancestry between the focal node and these individuals. So, given the Turing example configuration, the script will look to see if Richard Feynman and Alan Turing have a common ancestor, and if so, it will include both their ancestries in the genealogy. - Tree pruning (
CULL_AND_ABOVEandERASE_INDIVIDUAL): Including a particular academic can introduce a large ancestry and produce an ungainly genealogy. These two parameters (the cull list and the erasure list) allow us to prune the tree. Culling (CULL_AND_ABOVE) will remove an individual and his/her entire ancestry. Erasure (ERASE_INDIVIDUAL) will remove a particular individual but leave his/her ancestors untouched. Culling (as opposed to erasure) an individual will also insert an ellipsis above its children nodes to indicate that part of the tree was removed there. - Colour scheme: Colour individuals in the genealogy based on their relationship with the focal academic. This includes colouring based on whether the individual shares a common ancestor, is a direct ancestor, is a direct descendant, and so on. Colour instructions are included in the output dot file; most applications (e.g., graphviz and OmniGraffle) should be able to interpret these instructions.
You’ll likely need a bit of familiarity with Python syntax to get the most out of the script. Loading a Python module for configuration is a bit of a taboo, but is convenient enough for what this script needs to do. I’ve used networkx to make manipulating the genealogy (which is, more formally, a directed acyclic graph) simpler, since the script needs to handle joining and splitting of subgraphs, culling of disconnected components, and do some traversals for node colouring.
The script also takes a list of scientific prize winners. If any of these appear in the genealogy, they will be given a special colour, as per the colour scheme. Which prizes you wish to include is up to you. The Script-FindPrizeWinners directory contains a crude script that will compare the names of academics (which can be copy and pasted from a dot file for convenience) to prize winners and return any matches, so you can figure out who in your genealogy is a winner. I’ve included a few text files containing lists of winners (up to 2013) for various scientific prizes; namely, Abel, Cole, Fields, Turing, and Wolf prizes. It does fuzzy string matching, since the Wikipedia lists of winners (from where the names are sourced) might have slightly different spellings to those in the MGP, so it will likely produce false-positives – please use as a starting point only.
Design
After a few iterations of generating a dot file, checking its graphviz rendering, and tweaking the original configuration (e.g., adding more seeds, culling unwanted subtrees, erasing some nodes, etc.) I went on to import the file into OmniGraffle to do a prettier design. For anyone that wants somewhere to start, I’ve included the final design for one of my PhD supervisors, Roger Whitaker (who, interestingly, connects to my other supervisor, Stuart Allen, through William Hopkins), in the Designs directory. It’s in A-paper ratio ($1:\sqrt{2}$) but will need resizing to whatever print size is required. I had it printed on glossy A3 paper and put it in
this John Lewis picture frame.
(N.b.: Kudos to Stuart for kicking off this idea by stumbling on one of the older MGP genealogy scripts.)